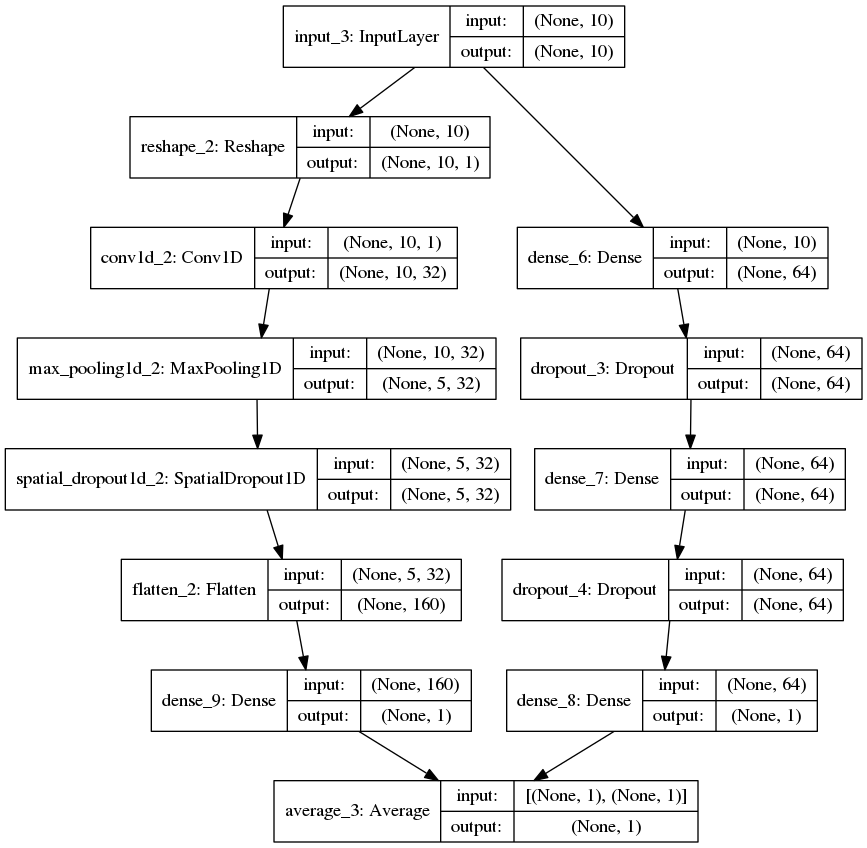
In the post (https://statcompute.wordpress.com/2017/01/08/an-example-of-merge-layer-in-keras), it was shown how to build a merge-layer DNN by using the Keras Sequential model. In the example below, I tried to scratch a merge-layer DNN with the Keras functional API in both R and Python. In particular, the merge-layer DNN is the average of a multilayer perceptron network and a 1D convolutional network, just for fun and curiosity. Since the purpose of this exercise is to explore the network structure and the use case of Keras API, I didn’t bother to mess around with parameters.
This file contains bidirectional Unicode text that may be interpreted or compiled differently than what appears below. To review, open the file in an editor that reveals hidden Unicode characters.
Learn more about bidirectional Unicode characters
| library(keras) | |
| df <- read.csv("credit_count.txt") | |
| Y <- matrix(df[df$CARDHLDR == 1, ]$DEFAULT) | |
| X <- scale(df[df$CARDHLDR == 1, ][3:14]) | |
| inputs <- layer_input(shape = c(ncol(X))) | |
| mlp <- inputs %>% | |
| layer_dense(units = 64, activation = 'relu', kernel_initializer = 'he_uniform') %>% | |
| layer_dropout(rate = 0.2, seed = 1) %>% | |
| layer_dense(units = 64, activation = 'relu', kernel_initializer = 'he_uniform') %>% | |
| layer_dropout(rate = 0.2, seed = 1) %>% | |
| layer_dense(1, activation = 'sigmoid') | |
| cnv <- inputs %>% | |
| layer_reshape(c(ncol(X), 1)) %>% | |
| layer_conv_1d(32, 4, activation = 'relu', padding = "same", kernel_initializer = 'he_uniform') %>% | |
| layer_max_pooling_1d(2) %>% | |
| layer_spatial_dropout_1d(0.2) %>% | |
| layer_flatten() %>% | |
| layer_dense(1, activation = 'sigmoid') | |
| avg <- layer_average(c(mlp, cnv)) | |
| mdl <- keras_model(inputs = inputs, outputs = avg) | |
| mdl %>% compile(optimizer = optimizer_sgd(lr = 0.1, momentum = 0.9), loss = 'binary_crossentropy', metrics = c('binary_accuracy')) | |
| mdl %>% fit(x = X, y = Y, epochs = 50, batch_size = 1000, verbose = 0) | |
| mdl %>% predict(x = X) |
This file contains bidirectional Unicode text that may be interpreted or compiled differently than what appears below. To review, open the file in an editor that reveals hidden Unicode characters.
Learn more about bidirectional Unicode characters
| from numpy.random import seed | |
| from pandas import read_csv, DataFrame | |
| from sklearn.preprocessing import scale | |
| from keras.layers.convolutional import Conv1D, MaxPooling1D | |
| from keras.layers.merge import average | |
| from keras.layers import Input, Dense, Flatten, Reshape, Dropout, SpatialDropout1D | |
| from keras.models import Model | |
| from keras.optimizers import SGD | |
| from keras.utils import plot_model | |
| df = read_csv("credit_count.txt") | |
| Y = df[df.CARDHLDR == 1].DEFAULT | |
| X = scale(df[df.CARDHLDR == 1].iloc[:, 2:12]) | |
| D = 0.2 | |
| S = 1 | |
| seed(S) | |
| ### INPUT DATA | |
| inputs = Input(shape = (X.shape[1],)) | |
| ### DEFINE A MULTILAYER PERCEPTRON NETWORK | |
| mlp_net = Dense(64, activation = 'relu', kernel_initializer = 'he_uniform')(inputs) | |
| mlp_net = Dropout(rate = D, seed = S)(mlp_net) | |
| mlp_net = Dense(64, activation = 'relu', kernel_initializer = 'he_uniform')(mlp_net) | |
| mlp_net = Dropout(rate = D, seed = S)(mlp_net) | |
| mlp_out = Dense(1, activation = 'sigmoid')(mlp_net) | |
| mlp_mdl = Model(inputs = inputs, outputs = mlp_out) | |
| ### DEFINE A CONVOLUTIONAL NETWORK | |
| cnv_net = Reshape((X.shape[1], 1))(inputs) | |
| cnv_net = Conv1D(32, 4, activation = 'relu', padding = "same", kernel_initializer = 'he_uniform')(cnv_net) | |
| cnv_net = MaxPooling1D(2)(cnv_net) | |
| cnv_net = SpatialDropout1D(D)(cnv_net) | |
| cnv_net = Flatten()(cnv_net) | |
| cnv_out = Dense(1, activation = 'sigmoid')(cnv_net) | |
| cnv_mdl = Model(inputs = inputs, outputs = cnv_out) | |
| ### COMBINE MLP AND CNV | |
| con_out = average([mlp_out, cnv_out]) | |
| con_mdl = Model(inputs = inputs, outputs = con_out) | |
| sgd = SGD(lr = 0.1, momentum = 0.9) | |
| con_mdl.compile(optimizer = sgd, loss = 'binary_crossentropy', metrics = ['binary_accuracy']) | |
| con_mdl.fit(X, Y, batch_size = 2000, epochs = 50, verbose = 0) | |
| plot_model(con_mdl, to_file = 'model.png', show_shapes = True, show_layer_names = True) |

You must be logged in to post a comment.