In my previous post https://statcompute.wordpress.com/2019/02/03/sobol-sequence-vs-uniform-random-in-hyper-parameter-optimization/, I’ve shown the difference between the uniform pseudo random and the quasi random number generators in the hyper-parameter optimization of machine learning.
Latin Hypercube Sampling (LHS) is another interesting way to generate near-random sequences with a very simple idea. Let’s assume that we’d like to perform LHS for 10 data points in the 1-dimension data space. We first partition the whole data space into 10 equal intervals and then randomly select a data point from each interval. For the N-dimension LHS with N > 1, we just need to independently repeat the 1-dimension LHS for N times and then randomly combine these sequences into a list of N-tuples.
LHS is similar to the Uniform Random in the sense that the Uniform Random number is drawn within each equal-space interval. On the other hand, LHS covers the data space more evenly in a way similar to the Quasi Random, such as Sobol Sequence. A comparison below shows how each of three looks like in the 2-dimension data space.
unifm_2d <- function(n, seed) {
set.seed(seed)
return(replicate(2, runif(n)))
}
sobol_2d <- function(n, seed) {
return(randtoolbox::sobol(n, dim = 2, scrambling = 3, seed = seed))
}
latin_2d <- function(n, seed) {
set.seed(seed)
return(lhs::randomLHS(n, k = 2))
}
par(mfrow = c(1, 3))
plot(latin_2d(100, 2019), main = "LATIN HYPERCUBE", xlab = '', ylab = '', cex = 2, col = "blue")
plot(sobol_2d(100, 2019), main = " SOBOL SEQUENCE", xlab = '', ylab = '', cex = 2, col = "red")
plot(unifm_2d(100, 2019), main = " UNIFORM RANDOM", xlab = '', ylab = '', cex = 2, col = "black")

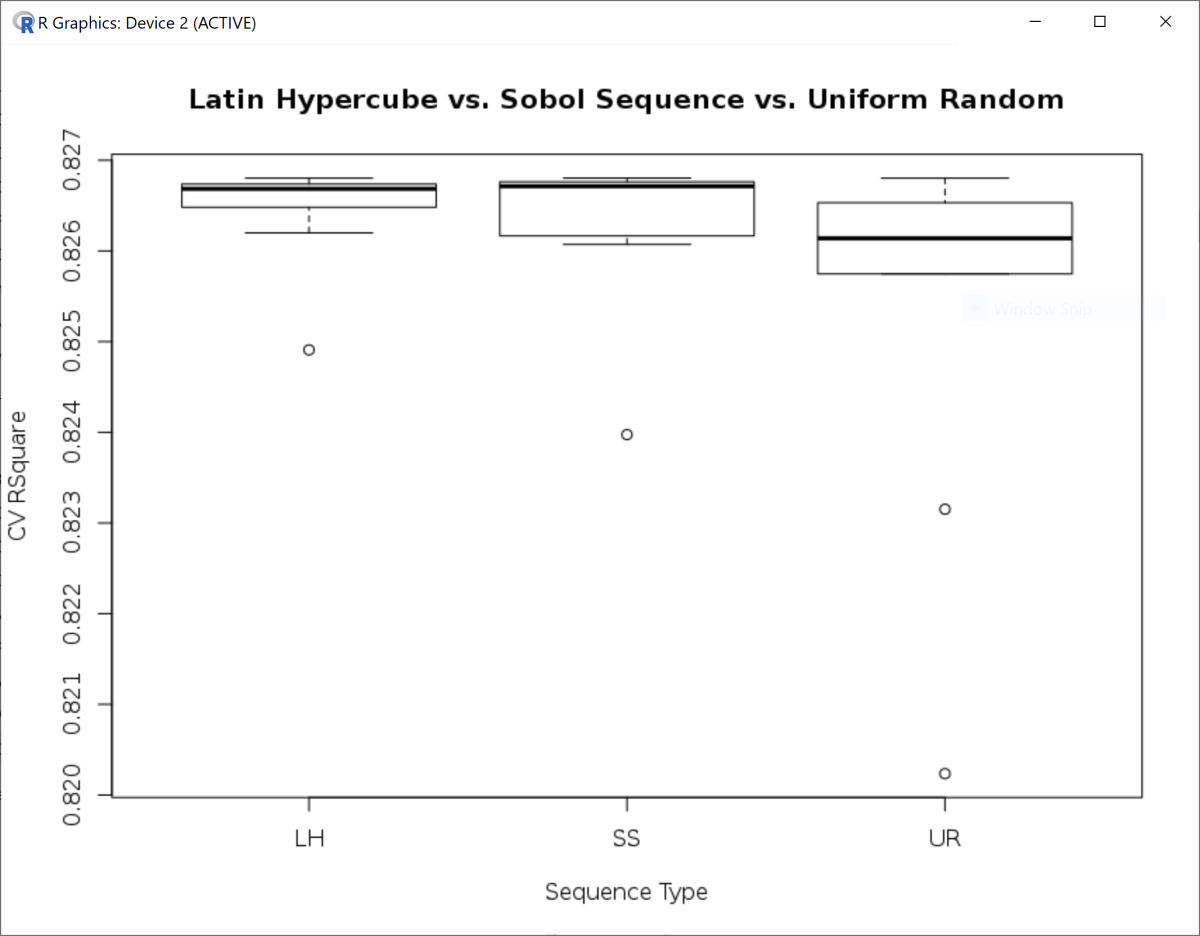
In the example below, three types of random numbers are applied to the hyper-parameter optimization of General Regression Neural Network (GRNN) in the 1-dimension case. While both Latin Hypercube and Sobol Sequence generate similar averages of CV R-squares, the variance of CV R-squares for Latin Hypercube is much lower. With no surprise, the performance of simple Uniform Random remains the lowest, e.g. lower mean and higher variance.
data(Boston, package = "MASS")
df <- data.frame(y = Boston[, 14], scale(Boston[, -14]))
gn <- grnn::smooth(grnn::learn(df), sigma = 1)
grnn.predict <- function(nn, dt) {
Reduce(c, lapply(seq(nrow(dt)), function(i) grnn::guess(nn, as.matrix(dt[i, ]))))
}
r2 <- function(act, pre) {
return(1 - sum((pre - act) ^ 2) / sum((act - mean(act)) ^ 2))
}
grnn.cv <- function(nn, sigmas, nfolds, seed = 2019) {
dt <- nn$set
set.seed(seed)
fd <- caret::createFolds(seq(nrow(dt)), k = nfolds)
cv <- function(s) {
rs <- Reduce(rbind,
lapply(fd,
function(f) data.frame(Ya = nn$Ya[unlist(f)],
Yp = grnn.predict(grnn::smooth(grnn::learn(nn$set[unlist(-f), ]), s),
nn$set[unlist(f), -1]))))
return(data.frame(sigma = s, R2 = r2(rs$Ya, rs$Yp)))
}
cl <- parallel::makeCluster(min(nfolds, parallel::detectCores() - 1), type = "PSOCK")
parallel::clusterExport(cl, c("fd", "nn", "grnn.predict", "r2"), envir = environment())
rq <- Reduce(rbind, parallel::parLapply(cl, sigmas, cv))
parallel::stopCluster(cl)
return(rq[rq$R2 == max(rq$R2), ])
}
gen_unifm <- function(min = 0, max = 1, n, seed) {
set.seed(seed)
return(round(min + (max - min) * runif(n), 8))
}
gen_sobol <- function(min = 0, max = 1, n, seed) {
return(round(min + (max - min) * randtoolbox::sobol(n, dim = 1, scrambling = 3, seed = seed), 8))
}
gen_latin <- function(min = 0, max = 1, n, seed) {
set.seed(seed)
return(round(min + (max - min) * c(lhs::randomLHS(n, k = 1)), 8))
}
nfold <- 10
nseed <- 10
sobol_out <- Reduce(rbind, lapply(seq(nseed), function(x) grnn.cv(gn, gen_sobol(0.1, 1, 10, x), nfold)))
latin_out <- Reduce(rbind, lapply(seq(nseed), function(x) grnn.cv(gn, gen_latin(0.1, 1, 10, x), nfold)))
unifm_out <- Reduce(rbind, lapply(seq(nseed), function(x) grnn.cv(gn, gen_unifm(0.1, 1, 10, x), nfold)))
out <- rbind(cbind(type = rep("LH", nseed), latin_out),
cbind(type = rep("SS", nseed), sobol_out),
cbind(type = rep("UR", nseed), unifm_out))
title <- "Latin Hypercube vs. Sobol Sequence vs. Uniform Random"
boxplot(R2 ~ type, data = out, main = title, ylab = "CV RSquare", xlab = "Sequence Type")
aggregate(R2 ~ type, data = out, function(x) round(c(avg = mean(x), var = var(x)), 8))
#type R2.avg R2.var
# LH 0.82645414 0.00000033
# SS 0.82632171 0.00000075
# UR 0.82536693 0.00000432










You must be logged in to post a comment.